
For the production of microfluidic operating systems and micropatterned surfaces for the investigation of biomolecules and cells, we develop tailor-made manufacturing processes and instruments. Examples include additive manufacturing processes (e.g. 3D printing), micro milling, or polymer pen lithography printers. We use CAM/CAD methods, fluidic simulations, micromechanical chip-to-world interfaces and machine-assisted liquid dispensing to manufacture the systems. Applications include biocatalysis, protein engineering and the investigation of biofilms and eukaryotic cells.
Selected References
- Arrabito, G., Schroeder, H., Schroder, K., Filips, C., Marggraf, U., Dopp, C., Venkatachalapathy, M., Dehmelt, L., Bastiaens, P. I., Neyer, A., Niemeyer, C. M. (2014) Configurable Low-Cost Plotter Device for Fabrication of Multi-Color Sub-Cellular Scale Microarrays. Small 10, 2870
- Hansen, S. H., Kabbeck, T., Radtke, C. P., Krause, S., Krolitzki, E., Peschke, T., Gasmi, J., Rabe, K. S., Wagner, M., Horn, H., Hubbuch, J., Gescher, J., Niemeyer, C. M. (2019) Machine-assisted cultivation and analysis of biofilms. Sci Rep 9, 8933.
- Grosche, M., Zoheir, A. E., Stegmaier, J., Mikut, R., Mager, D., Korvink, J. G., Rabe, K. S., Niemeyer, C. M. (2019) Microfluidic Chips for Life Sciences-A Comparison of Low Entry Manufacturing Technologies. Small 15, e1901956
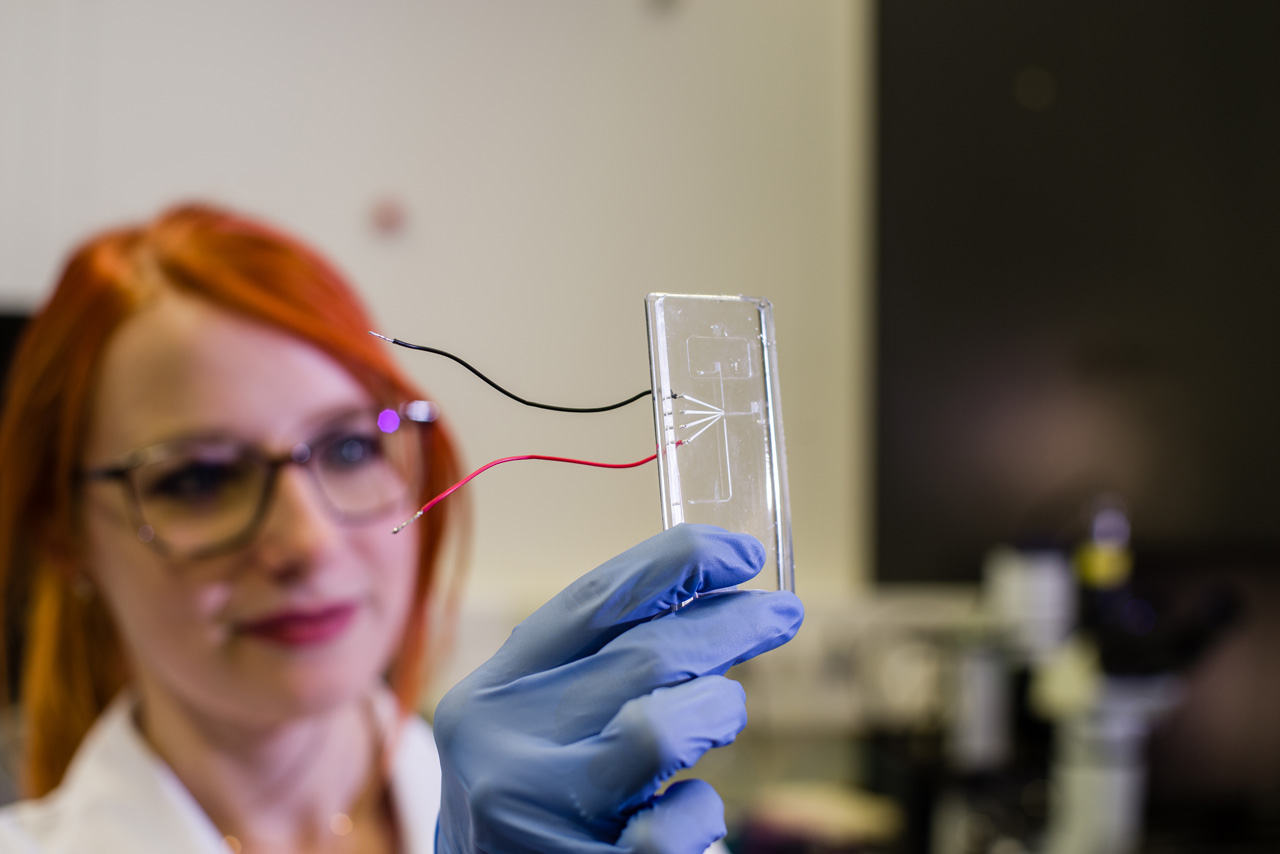
We develop customized microfluidic operating systems to produce biomolecules, to analyze biomolecular and cellular interactions and to study cell populations on synthetically modified surfaces. The fluidic systems, e.g. linear and branched channel systems, gradient mixers, microfluidic droplets and related accessories, are produced by modern manufacturing processes available in the institute. Application examples include microfluidic detection systems for diagnostics, bioreactors for the investigation of biofilms or for the characterization of cell-free biocatalysis processes.
Selected References
- Kampe, T., Konig, A., Schroeder, H., Hengstler, J. G., Niemeyer, C. M. (2014) Modular Microfluidic System for Emulation of Human Phase I/Phase II Metabolism. Anal Chem 86, 3068.
- Peschke, T., Skoupi, M., Burgahn, T., Gallus, S., Ahmed, I., Rabe, K. S., Niemeyer, C. M. (2017) Self-Immobilizing Fusion Enzymes for Compartmentalized Biocatalysis. ACS Catalysis 7, 7866
- Grösche, M., Korvink, J. G., Rabe, K. S., Niemeyer, C. M. (2019) Comparison of Storage Methods for Microfluidically Produced Water-in-Oil Droplets. Chem Engin Technol 42, DOI: 10.1002/ceat.201900075
- Hansen, S. H., Kabbeck, T., Radtke, C. P., Krause, S., Krolitzki, E., Peschke, T., Gasmi, J., Rabe, K. S., Wagner, M., Horn, H., Hubbuch, J., Gescher, J., Niemeyer, C. M. (2019) Machine-assisted cultivation and analysis of biofilms. Sci Rep 9, 8933.

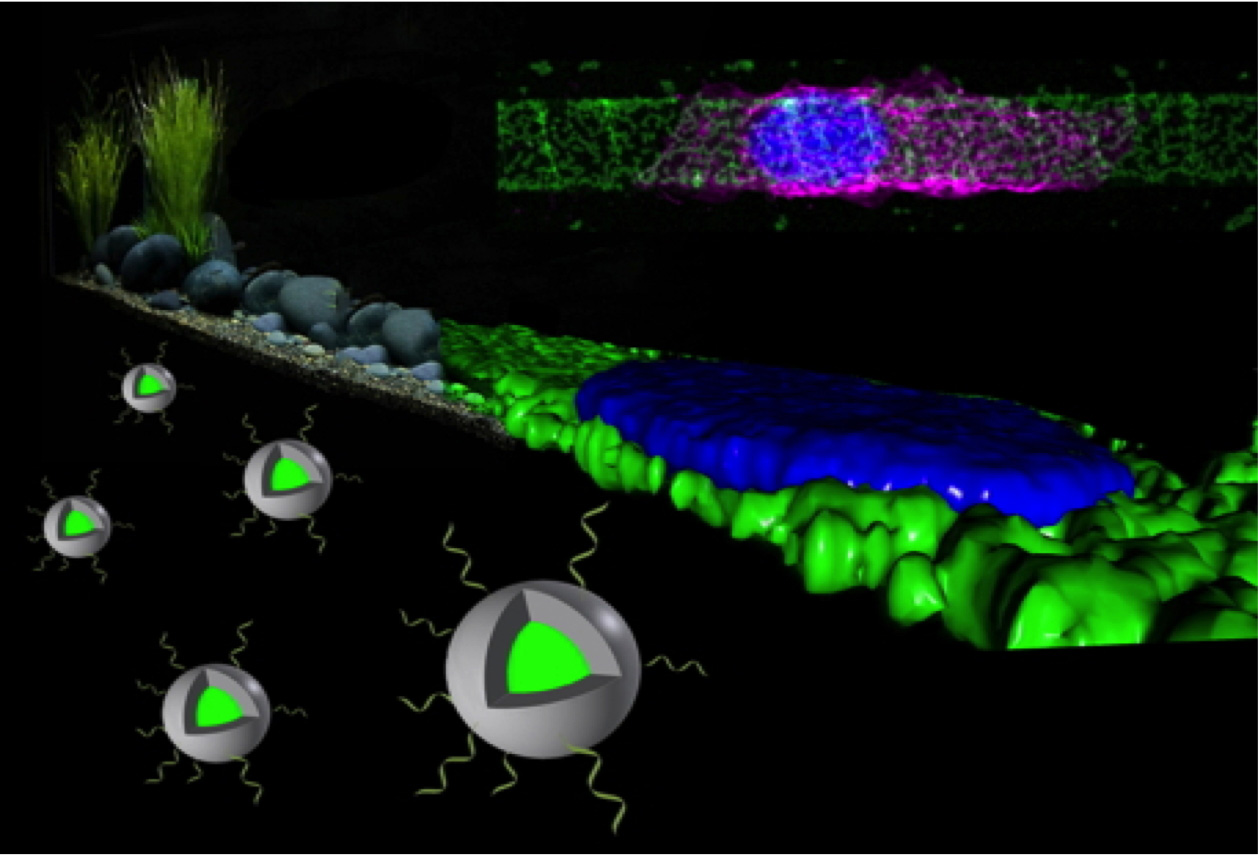
We have a long history of expertise in the development of DNA and protein microarrays used to study biomolecular interactions. Surface patterning is technically achieved by high-throughput micro-needle or ink-jet printing, micro contact printing, or polymer-pen lithography. Today, we mainly use biopatterned surfaces to study eukaryotic cells, their interaction with technical surfaces and the signalling cascades that occur within them. To this end, we have established a rich repertoire of methods ranging from surface chemistry and the production of biomolecular functional coatings to tailor-made operating systems for cultivation and coordinated high-resolution microscopy.
Selected References
- Arrabito, G., Reisewitz, S., Dehmelt, L., Bastiaens, P. I., Pignataro, B., Schroeder, H., Niemeyer, C. M. (2013) Biochips for Cell Biology by Combined Dip-Pen Nanolithography and DNA-Directed Protein Immobilization. Small 9, 4243
- Meyer, R., Sacca, B., Niemeyer, C. M. (2015) Site-Directed, On-Surface Assembly of DNA Nanostructures. Angew Chem Int Ed Engl 54, 12039
- Leidner, A., Weigel, S., Bauer, J., Reiber, J., Angelin, A., Grösche, M., Scharnweber, T., Niemeyer, C. M. (2018) Biopebbles: DNA‐Functionalized Core–Shell Silica Nanospheres for Cellular Uptake and Cell Guidance Studies. Funct. Mater. 28, 1707572
- Schneider, A.-K., Niemeyer, C. M. (2018) DNA Surface Technology: From Gene Sensors to Integrated Systems for Life and Materials Sciences. Angew Chem Int Ed 57, 16959-16967
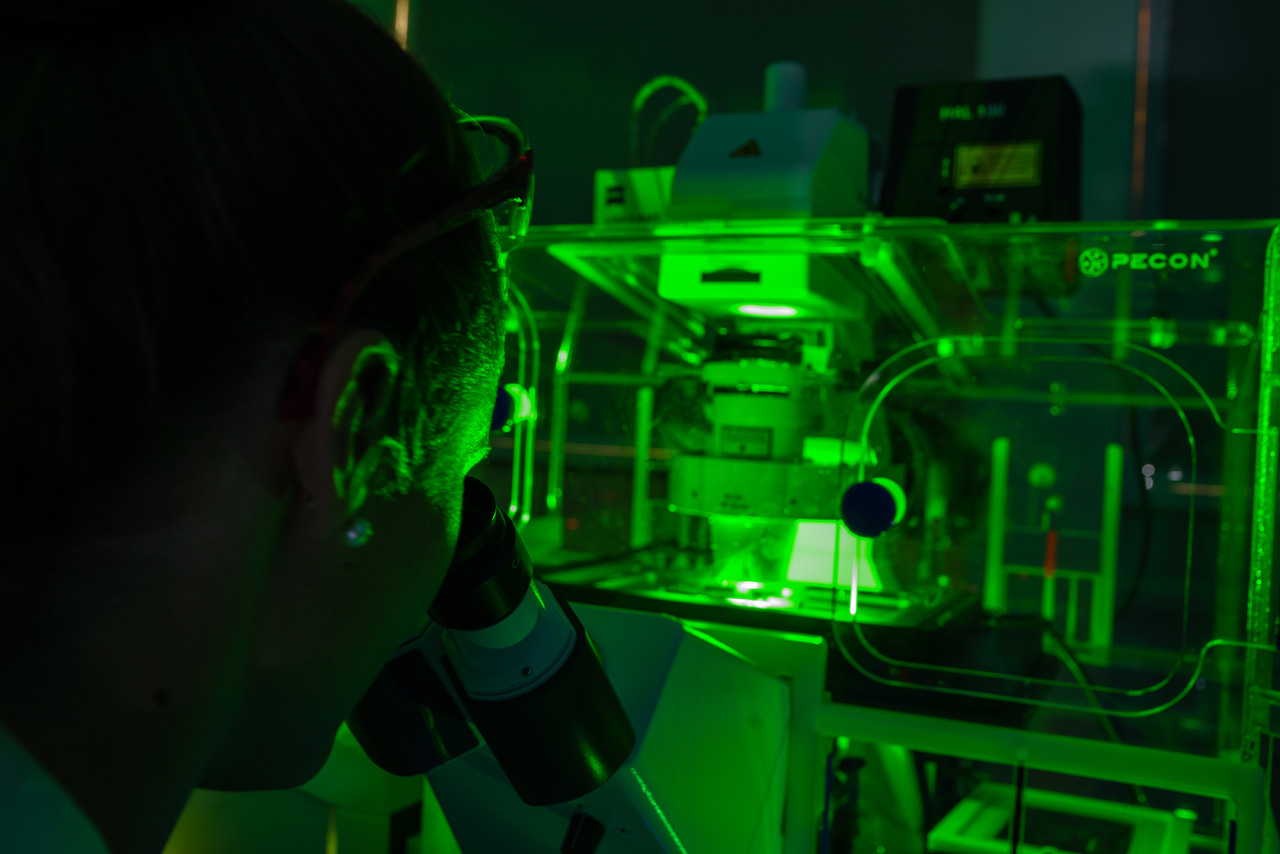
The institute is equipped with the latest generation of microscopes in order to be able to examine the micro- and nanostructured surfaces we have developed, the fluidic microsystems and the cell populations contained in these operating systems with maximum precision. These include confocal fluorescence microscopy, total-internal reflection (TIRF) fluorescence microscopy, high-speed fluorescence microscopy for dynamic systems, Keyance XXX system for characterization of microstructures, as well as two state-of-the-art atomic force microscopes for imaging nanostructures and force microscopy analyses of materials and cells.
Selected References
- Arrabito, G., Reisewitz, S., Dehmelt, L., Bastiaens, P. I., Pignataro, B., Schroeder, H., Niemeyer, C. M. (2013) Biochips for Cell Biology by Combined Dip-Pen Nanolithography and DNA-Directed Protein Immobilization. Small 9, 4243
- Meyer, R., Sacca, B., Niemeyer, C. M. (2015) Site-Directed, On-Surface Assembly of DNA Nanostructures. Angew Chem Int Ed Engl 54, 12039
- Leidner, A., Weigel, S., Bauer, J., Reiber, J., Angelin, A., Grösche, M., Scharnweber, T., Niemeyer, C. M. (2018) Biopebbles: DNA‐Functionalized Core–Shell Silica Nanospheres for Cellular Uptake and Cell Guidance Studies. Funct. Mater. 28, 1707572
- Schneider, A.-K., Niemeyer, C. M. (2018) DNA Surface Technology: From Gene Sensors to Integrated Systems for Life and Materials Sciences. Angew Chem Int Ed 57, 16959-16967

Analyzing, understanding and predicting biological systems is of key importance in order to substantially improve their precise engineering. The specific approaches are determined by the applications and include automated image recognition and analysis, force field calculations and molecular dynamics, multi-physics simulations, genetic analysis and machine learning approaches.
Selected References
- Buß,O., Muller, D., Jager, S., Rudat, J. and Rabe, K.S. (2018) Improvement in the Thermostability of a ß-Amino Acid Converting ω-Transaminase by Using FoldX. ChemBioChem 2018, 19, 379-387
- Grosche, M., Zoheir, A. E., Stegmaier, J., Mikut, R., Mager, D., Korvink, J. G., Rabe, K. S., Niemeyer, C. M. (2019) Microfluidic Chips for Life Sciences-A Comparison of Low Entry Manufacturing Technologies. Small 15, e1901956
- Li, G., Rabe, K. S., Nielsen, J., Engqvist, M. K. M., (2019) Machine learning applied to predicting microorganism growth temperatures and enzyme catalytic optima. ACS Synth. Biol. 8(6), 1411-1420

