
A key requirement for many approaches in our research is the availability of tailor-made proteins. Here, we employ a broad range of protein engineering strategies to provide the desired proteins in highly active configurations using bacterial and eukaryotic expression host systems to enable applications that range from mild and site-specific protein immobilization to biocatalysis. To optimize these systems, state-of-the-art directed evolution and bioinformatic approaches are being developed in the Molecular Evolution group headed by Kersten Rabe.
Selected References
- Koßmann, K. J., Ziegler, C., Angelin, A., Meyer, R., Skoupi, M., Rabe, K. S., Niemeyer, C. M. (2016) A rationally designed connector for assembly of protein-functionalized DNA nanostructures. ChemBioChem 17, 1102-1106
- Peschke, T., Rabe, K. S., Niemeyer, C. M. (2017) Orthogonal Surface Tags for Whole-Cell Biocatalysis. Angew Chem Int Ed Engl 56, 2183-2186
- Maier, M., Radtke, C.P., Hubbuch, J., Niemeyer, C.M. and Rabe K.S. (2018) On-Demand production of flow reactor cartridges by 3D printing of thermostable enzymes. Angew. Chem. Int. Ed. 57, 5539-5543
- Buß,O., Muller, D., Jager, S., Rudat, J. and Rabe, K.S. (2018) Improvement in the Thermostability of a ß-Amino Acid Converting ω-Transaminase by Using FoldX. ChemBioChem 2018, 19, 379-387
- Peschke, T., Bitterwolf, P., Gallus, S., Hu, Y., Oelschlaeger, C., Willenbacher, N., Rabe, K. S., Niemeyer, C. M. (2018) Self-Assembling All-Enzyme Hydrogels for Flow Biocatalysis. Angew Chem Int Ed 57, 17028-17032

From the outset, our research program has been geared towards making a contribution in the field of nanobiotechnology. To this end, we are developing strategies for the bottom-up assembly of nucleic acid nanostructures, such as DNA origami, and to functionalize them with proteins and nanoparticles to enable applications in the fields of biosensing, biocatalysis and cell biology.
Selected References
- Sacca, B., Meyer, R., Erkelenz, M., Kiko, K., Arndt, A., Schroeder, H., Rabe, K. S., Niemeyer, C. M. (2010) Orthogonal Protein Decoration of DNA Origami. Angew Chem Int Ed 49, 9378-9383; Angew. Chem. 122, 9568 -9573.
- Erkelenz, M., Kuo, C. H., Niemeyer, C. M. (2011) DNA-mediated assembly of Cytochrome P450 BM3 subdomains. J. Am. Chem. Soc. 133, 16111-16118
- Timm, C., Niemeyer, C. M. (2015) Assembly and Purification of Enzyme-Functionalized DNA Origami Structures. Angew Chem Int Ed Engl 54, 6745-6750
- Angelin, A., Weigel, S., Garrecht, R., Meyer, R., Bauer, J., Kumar, R. K., Hirtz, M., Niemeyer, C. M. (2015) Multiscale Origami Structures as Interface for Cells. Angew Chem Int Ed Engl 54, 15813
- Burgahn, T., Garrecht, R., Rabe, K. S., Niemeyer, C. M. (2019) Solid-Phase Synthesis and Purification of Protein–DNA Origami Nanostructures. Chem. Eur. J. 25, 3483
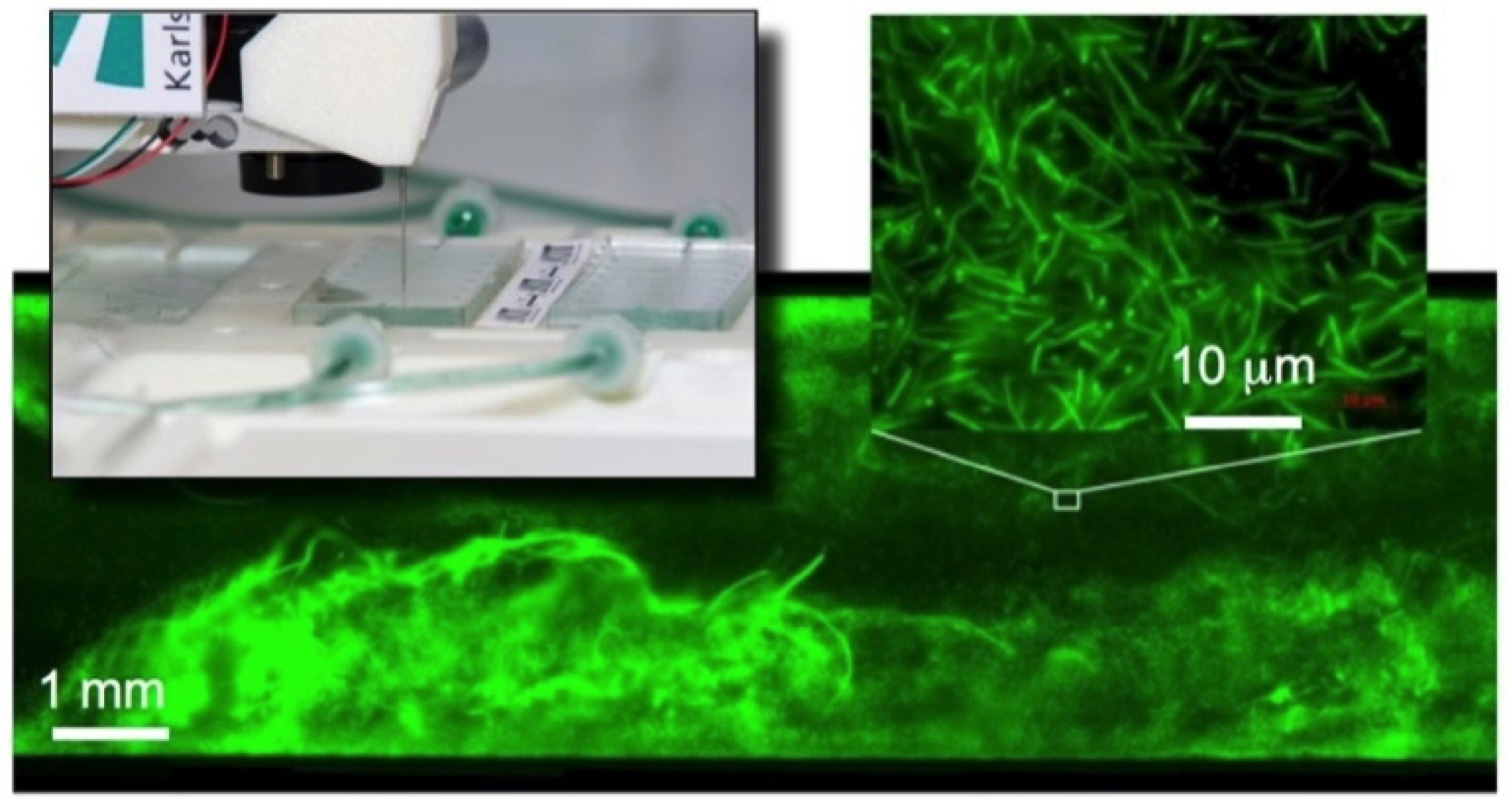
Biofilms are the natural life form of most microorganisms. These multispecies consortia are characterised by outstanding vitality and robustness, which leads to a variety of problems for technology and medicine, but also offers enormous potential for biotechnological processes.
In cooperation with the labs of Johannes Gescher and Harald Horn, we develop engineering solutions that enable the integrated, machine-assisted cultivation and analysis of biofilms in microfluidic operating systems. The aim of this work is to develop productive biofilms for industrial biotechnology.
Selected References
- Hansen, S. H., Kabbeck, T., Radtke, C. P., Krause, S., Krolitzki, E., Peschke, T., Gasmi, J., Rabe, K. S., Wagner, M., Horn, H., Hubbuch, J., Gescher, J., Niemeyer, C. M. (2019) Machine-assisted cultivation and analysis of biofilms. Sci Rep 9, 8933.
- Peschke, T., Bitterwolf, P., Hansen, S., Gasmi, J., Rabe, K. S., Niemeyer, C. M. (2019) Self-Immobilizing Biocatalysts Maximize Space–Time Yields in Flow Reactors. Catalysts 9, 164

We use our methodological repertoire for the production of bioconjugates and biopatterned surfaces to study the interaction of eukaryotic cells with solid matter and to exploit these interactions to study signal transduction processes in living cells. For this purpose, we use high-resolution fluorescence microscopy equipment of the latest generation, which enables us to study living cells in microfluidic cultivation chambers under native conditions.
Selected References
- Schroeder, H., Ellinger, B., Becker, C. F. W., Waldmann, H., Niemeyer, C. M. (2007) Generation of Live Cell Microarrays by Means of DNA-Directed Immobilization of Specific Cell Surface Ligands. Angew. Chem. Int. Ed. 46, 4180.
- Reisewitz, S., Schroeder, H., Tort, N., Edwards, K. A., Baeumner, A. J., Niemeyer, C. M. (2010) Capture and Culturing of Living Cells on Microstructured DNA Substrates. Small 6, 2162
- Gandor, S., Reisewitz, S., Venkatachalapathy, M., Arrabito, G., Reibner, M., Schroder, H., Ruf, K., Niemeyer, C. M., Bastiaens, P. I., Dehmelt, L. (2013) A Protein-Interaction Array Inside a Living Cell. Angew. Chem. Int. Ed. Engl. 52, 4890
- Angelin, A., Weigel, S., Garrecht, R., Meyer, R., Bauer, J., Kumar, R. K., Hirtz, M., Niemeyer, C. M. (2015) Multiscale Origami Structures as Interface for Cells. Angew Chem Int Ed Engl 54, 15813

